[ad_1]
Schistosomiasis is a disease of global medical and veterinary importance, caused by parasitic flatworms of the genus Schistosoma. Around 200 million people, and an untold number (possibly in the billions) of animals are estimated to be infected, globally.
Certain schistosome species can infect humans (e.g. Schistosoma mansoni and S. haematobium), others animals (S. bovis, S. curassoni), and some species can infect both humans and animals (i.e. are zoonotic, e.g. S. japonicum).
Schistosome hybrids
It is increasingly being observed that closely related schistosome species can interbreed, exchange genetic material, and produce viable offspring (i.e. hybridise) with one another, further complicating the transmission landscape. This is in addition to taxonomic species classification and control strategy implementation.
For example, within the S. haematobium group of 9 African species, hybrids have been found between nearly all major group members, such as S. haematobium and S. bovis, S. curassoni, and S. mattheei, and S. curassoni and S. bovis.
In addition, hybridisation events have the possibility of producing schistosome hybrids with increased virulence and parasite ‘fitness’ (i.e. hybrid vigour), and may lead to the rapid evolution of new zoonotic species. For example, hybridisation of a human-specific and animal-specific schistosome species (such as S. haematobium and S. bovis) may increase the host range of the resulting offspring, allowing the hybrid to infect both humans and animals.
Diagnostics developed to identify schistosome species, which forms a key component of any schistosomiasis surveillance program, may be rendered less effective through their inability to accurately detect these hybrids, thus presenting considerable challenges for control and prevention of the disease.
Therefore, rapid and low-cost diagnostics methods that can reliably identify schistosome hybrids are vital and urgently needed. Molecular diagnostic methods that detect parasite DNA (e.g. PCR) in patient or environmental samples fulfill these criteria, although require the detection of species-specific DNA sequences from both the nuclear (e.g. ITS and 18S) and mitochondrial (e.g. Cox1) genes of hybrid parasites.
Both nuclear and mitochondrial genes need to be used to detect hybrids, as the hybrid offspring inherits one of these from each of their parent species, such as the nuclear genome from S. haematobium, and the mitochondrial genome from S. bovis.
These particular genes are traditionally used for species identification because different schistosome species accrue mutations (i.e. single nucleotide polymorphisms, SNPs) in different locations on the genes, and so through identification (i.e. genotyping) of these SNPs, one can determine which schistosome species is present. Classically, molecular identification of each of these genes required different PCR protocols, raising the time and cost involved in schistosome species, and hybrid, identification.
Developing a new diagnostic method for schistosome species and hybrid incrimination
A group of collaborating researchers from France (University of Perpignan), the UK (Natural History Museum), Switzerland (University of Basel), Mali (University of Sciences, Techniques and Technologies of Bamako) and Côte d’Ivoire (Université Félix Houphouët-Boigny) have recently designed a new SNP genotyping molecular diagnostic assay able to detect three schistosome species (S. haematobium, S. bovis, S. curassoni), as well as their hybrid forms, in a single reaction; catchily called a duplex tetra-primer amplification refractory mutation system polymerase chain reaction (T-ARMS-PCR) assay.
There’s a lot to unpack in ‘duplex T-ARMS-PCR’, which combines principals of a tetra-primer PCR and an ARMS-PCR.
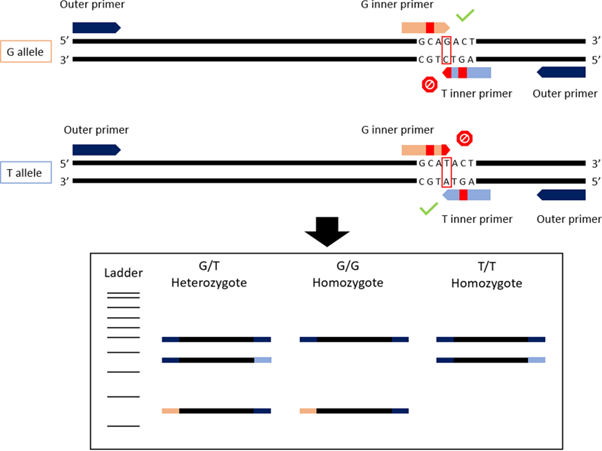
The ARMS-PCR aspect of the technique consists of SNP genotyping, where two of the primers bind to the gene region with a known SNP, or two different SNPs, where one SNP is characteristic of one species, and the other of a different schistosome species. Combining two ARMS-PCR methods together, each requiring the use of four primers (2 primer pairs) to amplify the target ITS and 18S gene regions, respectively, the ‘duplex’ tetra-primer ARMS-PCR is formed.
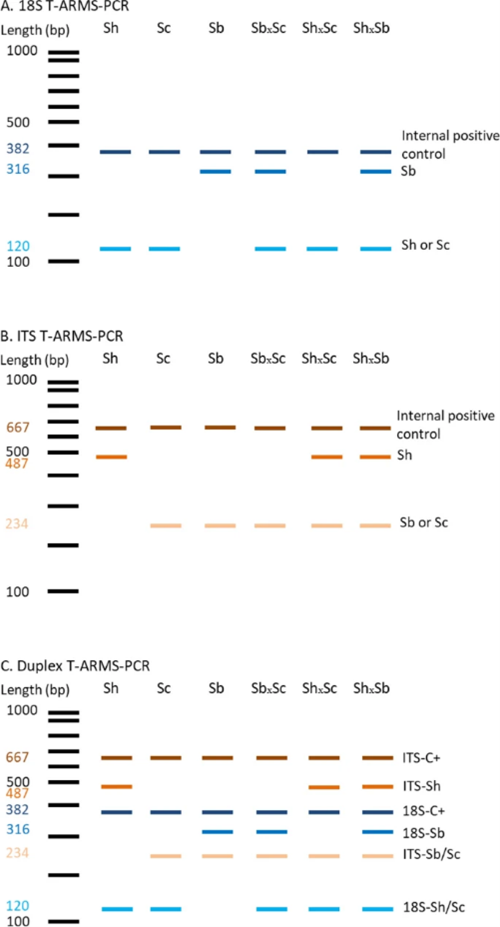
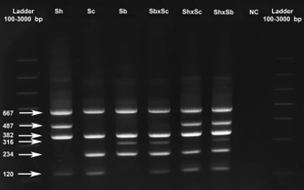
Each of the primer pairs results in amplification of 2 gene fragments of different lengths (and thus sizes) for the ITS and 18S targets, which can then be visualised using gel electrophoresis. The ITS-targeting PCR primers could separate S. bovis and S. curassoni from S. haematobium, as the two former species produce fragment sizes during gel electrophoresis (i.e. bands) of 243bp, and S. haematobium produces a 487bp band. The 18S-targeting PCR primers, on the other hand, distinguished between S. bovis (316bp band) and S. curassoni and S. haematobium (120bp band).
Ignoring the internal positive control (C+, bands 667bp and 382bp), if two bands are seen on the gel then this is indicative of a specific schistosome species, and if 3 bands are observed then the DNA of a hybrid schistosome has been detected. Therefore, combining both ITS and 18S ARMS-PCR methods into a single diagnostic tool, the researchers could distinguish between all three schistosome species, and their hybrids.
Results and implications
To test their method, the researchers used 169 laboratory and field-collected larval (miracidium) and adult parasite stages of the three target schistosome species from Spain, Egypt, Mali, Senegal and Côte d’Ivoire. Further to this, they validated their method by sequencing the ITS, 18S and Cox1 gene regions from 148 of the samples to confirm the schistosome species present, and whether it was a hybrid.
Assay testing and sequencing validation results show their diagnostic assay was able to detect all three schistosome species and their hybrids, with 100% specificity and sensitivity, in all 148 samples.
Despite this impressive result, the authors note that the assay could be further improved by the inclusion of mitochondrial markers to increase genotyping precision. Otherwise, the assay would require deployment alongside an existing mitochondrial genotyping diagnostic for complete hybrid identification.
Nonetheless, the development of this effective T-ARMS-PCR diagnostic assay, combining SNP genotyping protocols for two different nuclear gene markers, provides a single assay that’s fast, accurate, and relatively cheap.
The development and deployment of such diagnostics is crucial given the ever-changing schistosomiasis transmission landscape, and will allow the improved investigation of schistosome epidemiology, population genetics, and hybridisation events.
[ad_2]
Source link